Example JSim model generated from Modular Program Constructor (MPC). Three species (A, B, C), two compartment model with two reactions in compartment two with species concentrations described by ordinary differential equations (ODEs). Taken from 2015 F1000 article.
Description
This is the example used to reproduce Figure 2 (Solution to two compartment model generated from MPC.) of Jardine et al., 2015 paper describing the MPC software. To run simulation go to the 'Run Time' tab and hit the 'Run' button near the top of page. 'Figure2' plot page on left should display results as decribed in paper. External imput for species 'A' is an Lag Normal input function. There is no flow in of species 'B' and 'C'.
Model generation by MPC:
Please see MPC GitHub repository for MPC software and introduction. More information on MPC can also be found here. This model was created using two files:
a module library
and an MPC file
which specifies how MPC is to create the final model. These files are in plain text and you are encouraged to view them. They are annotated to assist in reviewing. The actual syntax to generate the model is: 'java -jar MPC.jar F1000_example.mpc', which creates a model (F1000_example.mod) that can be used by JSim.
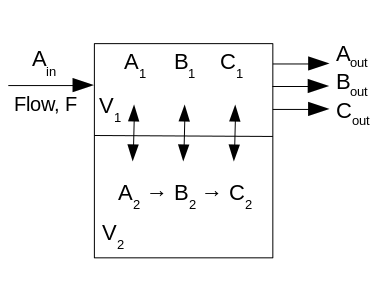
Figure 1: Two compartment, three species model (A, B, C) with volumes V1, V2, respectively. Ain is the concentration of A entering compartment 1 through which the flow is F. There is no flow in V2, but there are the reactions A-->B and B-->C. Passive exchange between compartments occurs for all three species.
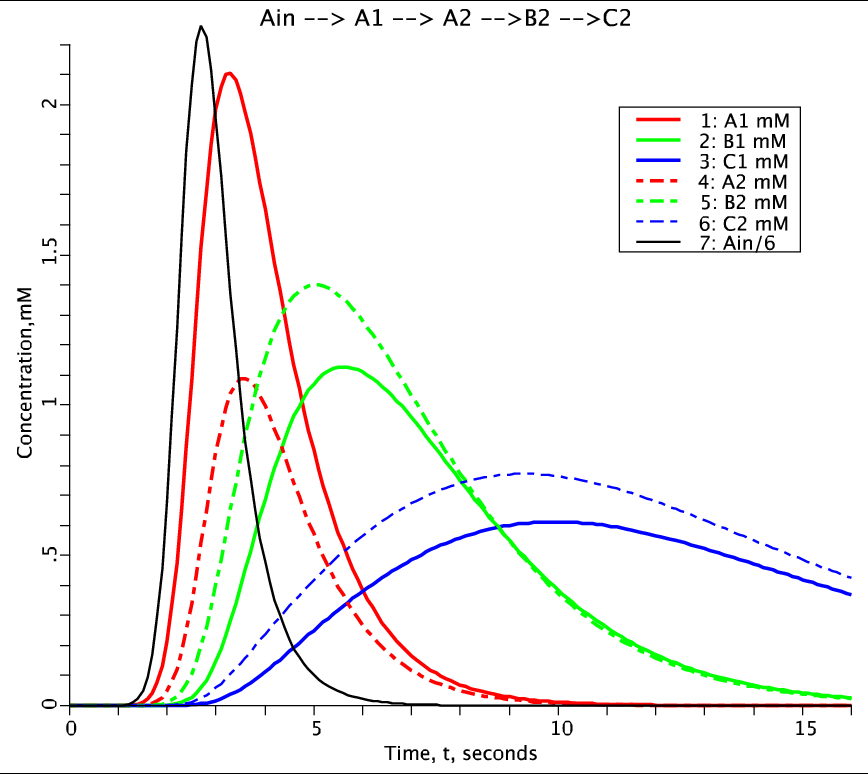
Figure 2: Solution to two compartment model generated from MPC. Species concentrations plotted as a function of time. Species A (red), B (green), C (blue). Compartment 1: solid line, Compartment 2: dashed line. Lag Normal external input for species A is Ain, the black solid line.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Jardine B, Raymond G and Bassingthwaighte J. Semi-automated Modular Program Constructor for physiological modeling: Building cell and organ models, [version 1; referees: awaiting peer review] F1000Research 2015, 4:1461 (doi: 10.12688/f1000research.7476.1) Butterworth E, Jardine BE, Raymond GM, Neal ML, Bassingthwaighte JB. JSim, an open-source modeling system for data analysis, [v3; ref status: indexed, http://f1000r.es/3n0] F1000Research 2014, 2:288 (doi: 10.12688/f1000research.2-288.v3)
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

